Congratulations to Ed Braun, publishing related manuscripts in both Nature [https://www.nature.com/articles/s41586-024-07323-1] and PNAS [https://doi.org/10.1073/pnas.2319506121] on the same day – April 1st!
Ed’s PNAS paper is entitled “A region of suppressed recombination misleads neoavian phylogenomics”. Ed and his colleagues found that it is possible use computational methods to detect a strong signal of chromosomal rearrangements that occurred about 65 million years ago in birds – near the time when dinosaurs went extinct and most modern birds appeared.
Ed’s Nature paper: “Complexity of avian evolution revealed by family-level genomes” is part of the Bird 10,000 Genomes (B10K) consortium tasked with sequencing the genomes of every single species of bird. With this data the team created the most detailed bird evolutionary tree, detailing how bird groups diversified after the extinction of dinosaurs.
PNAS paper news:
https://www.eurekalert.org/news-releases/1039323.
https://phys.org/news/2024-04-tools-fuel-reconstruction-bird-family.html.
https://www.popsci.com/science/bird-evolution-wrong/
https://www.earth.com/news/untold-story-of-bird-evolution-revealed-through-genetics/
For a detailed summary of the PNAS paper see below:
- It has been difficult to infer relationships among the major groups of birds, even using data from complete bird genomes.
- We found that whole genome analyses support two different answers depending on the number of species included in the analyses:
- Analyses using many representatives of most major groups support splitting Neoaves (most bird species) into two major groups: 1) Flamingos + Grebes; and 2) other Neoaves
- Analyses using a few representatives of each major group support splitting Neoaves into two different groups: 1) Doves (and two less well-known groups) + Flamingos + Grebes; and 2) other Neoaves
- This conflict is driven by a region of about 20 million DNA base pairs on one single chromosome (birds have about 1 billion DNA base pairs in their genomes). Genes in the region strongly support Doves + Flamingos + Grebes vs Others whereas genes spread across the genome support Flamingos + Grebes vs Others (where others include Doves)
- This can be explained by know molecular biology – crossover during reprodution were suppressed on the “outlier” chromosome for a period of about 2-3 million years due to rearranged DNA
- The period of suppressed crossovers occurred about 65 million years ago.
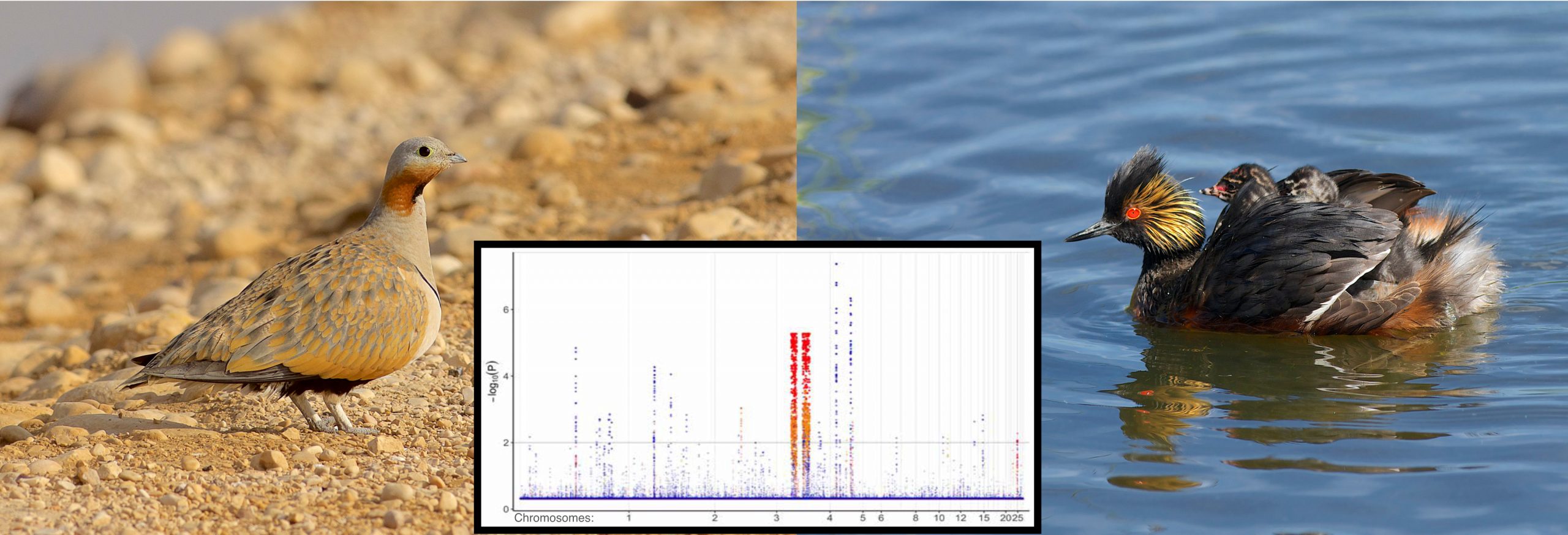
The inset graph shows two anomalous regions that map to chromosome 4 of the chicken genome where there is strong signal uniting Mirandornithes (flamingos and grebes,) and Columbimorphae (doves, mesites, and sandgrouse). Mirandornithes is not closely related to the Columbimorphae for the rest of the genome. This unexpected distribution of signal reflects suppressed recombination for a period of approximately 3 million years more than 60 million years ago.
